Redun¶
Here, we’ll see how to track redun workflow runs with LaminDB.
Note
This use case is based on github.com/ricomnl/bioinformatics-pipeline-tutorial.
# pip install lamindb redun git+http://github.com/laminlabs/redun-lamin-fasta
!lamin init --storage ./test-redun-lamin
Show code cell output
→ initialized lamindb: testuser1/test-redun-lamin
Amend the workflow¶
import lamindb as ln
import json
Show code cell output
→ connected lamindb: testuser1/test-redun-lamin
Let’s amend a redun workflow.py to register input & output artifacts in LaminDB:
To track the workflow run in LaminDB, add (see on GitHub):
ln.track(params=params)
To register the output file via LaminDB, add (see on GitHub):
ln.Artifact(output_path, description="results").save()
Run redun¶
Let’s see what the input files are:
!ls ./fasta
Show code cell output
KLF4.fasta MYC.fasta PO5F1.fasta SOX2.fasta
And call the workflow:
!redun run workflow.py main --input-dir ./fasta --tag run=test-run 1> run_logs.txt 2>run_logs.txt
Inspect the logs:
!cat run_logs.txt
Show code cell output
→ connected lamindb: testuser1/test-redun-lamin
→ created Transform('fncb7ZrLZP3V0000'), started new Run('ZKnkCpA9...') at 2025-06-10 20:14:56 UTC
→ params: input_dir=./fasta, amino_acid=C, enzyme_regex=[KR], missed_cleavages=0, min_length=4, max_length=75, executor=Executor.default
• recommendation: to identify the script across renames, pass the uid: ln.track("fncb7ZrLZP3V", params={...})
! folder i[redun] Run Job d70d26d2: bioinformatics_pipeline_tutorial.lib.digest_protein_task(input_fasta=File(path=/home/runner/work/redun-lamin/→ finished Run('ZKnkCpA9') after 6s at 2025-06-10 20:15:03 UTC
File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-lamin/data/results.tgz, hash=0b79e77e)
Job 0a2aaade: bioinformatics_pipeline_tutorial.lib.digest_protein_task(input_fasta=File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-lamin/.lamindb/UOFY7yU4ekvGM6i40000.fasta, hash=05f36b7f), enzyme_regex='[KR]', missed_cleavages=0, min_length=4, max_length=75) on default
[redun] Run Job 8a732b2c: bioinformatics_pipeline_tutorial.lib.digest_protein_task(input_fasta=File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-lamin/.lamindb/DpeUKSeDgo5KxsYq0000.fasta, hash=edbccebd), enzyme_regex='[KR]', missed_cleavages=0, min_length=4, max_length=75) on default
[redun] Run Job a0869329: bioinformatics_pipeline_tutorial.lib.digest_protein_task(input_fasta=File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-lamin/.lamindb/mdMdoSp7KpKk5EW60000.fasta, hash=8d7ae32e), enzyme_regex='[KR]', missed_cleavages=0, min_length=4, max_length=75) on default
[redun] Run Job 41279a9a: bioinformatics_pipeline_tutorial.lib.count_amino_acids_task(input_fasta=File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-lamin/.lamindb/l5zDAPj3F6hQm21j0000.fasta, hash=58129eeb), input_peptides=File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-lamin/data/l5zDAPj3F6hQm21j0000.peptides.txt, hash=d2e94ce9), amino_acid='C') on default
[redun] Run Job 8f952768: bioinformatics_pipeline_tutorial.lib.count_amino_acids_task(input_fasta=File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-lamin/.lamindb/UOFY7yU4ekvGM6i40000.fasta, hash=05f36b7f), input_peptides=File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-lamin/data/UOFY7yU4ekvGM6i40000.peptides.txt, hash=5ba81227), amino_acid='C') on default
[redun] Run Job 310ac865: bioinformatics_pipeline_tutorial.lib.count_amino_acids_task(input_fasta=File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-lamin/.lamindb/DpeUKSeDgo5KxsYq0000.fasta, hash=edbccebd), input_peptides=File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-lamin/data/DpeUKSeDgo5KxsYq0000.peptides.txt, hash=34cf438f), amino_acid='C') on default
[redun] Run Job cd85f412: bioinformatics_pipeline_tutorial.lib.count_amino_acids_task(input_fasta=File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-lamin/.lamindb/mdMdoSp7KpKk5EW60000.fasta, hash=8d7ae32e), input_peptides=File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-lamin/data/mdMdoSp7KpKk5EW60000.peptides.txt, hash=27458788), amino_acid='C') on default
[redun] Run Job e8df22a2: bioinformatics_pipeline_tutorial.lib.plot_count_task(input_count=File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-lamin/data/l5zDAPj3F6hQm21j0000.count.tsv, hash=623fba4d)) on default
[redun] Run Job 9390fca7: bioinformatics_pipeline_tutorial.lib.plot_count_task(input_count=File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-lamin/data/UOFY7yU4ekvGM6i40000.count.tsv, hash=50de614b)) on default
[redun] Run Job 8d8c032c: bioinformatics_pipeline_tutorial.lib.plot_count_task(input_count=File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-lamin/data/DpeUKSeDgo5KxsYq0000.count.tsv, hash=24892a75)) on default
[redun] Run Job 2dc4153d: bioinformatics_pipeline_tutorial.lib.plot_count_task(input_count=File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-lamin/data/mdMdoSp7KpKk5EW60000.count.tsv, hash=b1e7da18)) on default
[redun] Run Job 57a10ed8: bioinformatics_pipeline_tutorial.lib.get_report_task(input_counts=[File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-lamin/data/l5zDAPj3F6hQm21j0000.count.tsv, hash=623fba4d), File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-l...) on default
[redun] Run Job 42002643: bioinformatics_pipeline_tutorial.lib.archive_results_task(inputs_plots=[File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-lamin/data/l5zDAPj3F6hQm21j0000.plot.png, hash=a709338a), File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-la..., input_report=File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-lamin/data/protein_report.tsv, hash=0fb19e68)) on default
[redun] Run Job 6d581129: redun_lamin_fasta.finish(results_archive=File(path=/home/runner/work/redun-lamin/redun-lamin/docs/test-redun-lamin/data/results.tgz, hash=0b79e77e)) on default
[redun]
[redun] | JOB STATUS 2025/06/10 20:15:03
[redun] | TASK PENDING RUNNING FAILED CACHED DONE TOTAL
[redun] |
[redun] | ALL 0 0 0 0 16 16
[redun] | bioinformatics_pipeline_tutorial.lib.archive_results_task 0 0 0 0 1 1
[redun] | bioinformatics_pipeline_tutorial.lib.count_amino_acids_task 0 0 0 0 4 4
[redun] | bioinformatics_pipeline_tutorial.lib.digest_protein_task 0 0 0 0 4 4
[redun] | bioinformatics_pipeline_tutorial.lib.get_report_task 0 0 0 0 1 1
[redun] | bioinformatics_pipeline_tutorial.lib.plot_count_task 0 0 0 0 4 4
[redun] | redun_lamin_fasta.finish 0 0 0 0 1 1
[redun] | redun_lamin_fasta.main 0 0 0 0 1 1
[redun]
[redun]
[redun] Execution duration: 7.17 seconds
View data lineage:
artifact = ln.Artifact.get(key="data/results.tgz")
artifact.view_lineage()
Show code cell output
Track the redun execution id¶
If we want to be able to query LaminDB for redun execution ID, this here is a way to get it:
# export the run information from redun
!redun log --exec --exec-tag run=test-run --format json --no-pager > redun_exec.json
# load the redun execution id from the JSON and store it in the LaminDB run record
with open("redun_exec.json") as file:
redun_exec = json.loads(file.readline())
artifact.run.reference = redun_exec["id"]
artifact.run.reference_type = "redun_id"
artifact.run.save()
Run(uid='ZKnkCpA9XyR22zNM', started_at=2025-06-10 20:14:56 UTC, finished_at=2025-06-10 20:15:03 UTC, reference='31972db9-19bb-4a1c-a7c5-419180e73913', reference_type='redun_id', branch_id=1, space_id=1, transform_id=1, report_id=7, environment_id=6, created_by_id=1, created_at=2025-06-10 20:14:56 UTC)
Track the redun run report¶
Attach a run report:
report = ln.Artifact(
"run_logs.txt",
description=f"Redun run report of {redun_exec['id']}",
run=False,
visibility=0,
).save()
artifact.run.report = report
artifact.run.save()
Run(uid='ZKnkCpA9XyR22zNM', started_at=2025-06-10 20:14:56 UTC, finished_at=2025-06-10 20:15:03 UTC, reference='31972db9-19bb-4a1c-a7c5-419180e73913', reference_type='redun_id', branch_id=1, space_id=1, transform_id=1, report_id=8, environment_id=6, created_by_id=1, created_at=2025-06-10 20:14:56 UTC)
View transforms and runs in LaminHub¶
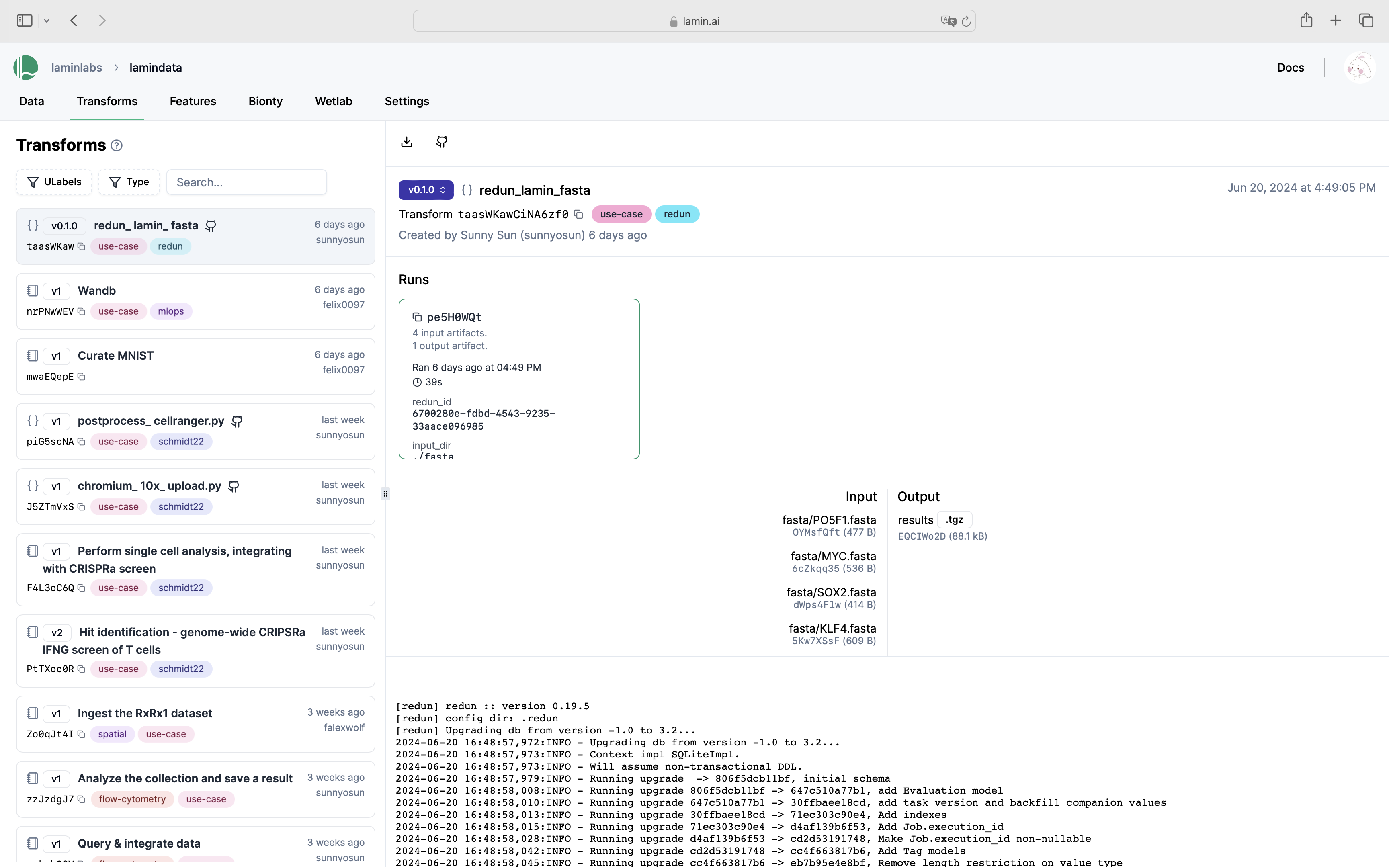
View the database content¶
ln.view()
Show code cell output
Artifact
| uid | key | description | suffix | kind | otype | size | hash | n_files | n_observations | _hash_type | _key_is_virtual | _overwrite_versions | space_id | storage_id | schema_id | version | is_latest | run_id | created_at | created_by_id | _aux | branch_id | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | |||||||||||||||||||||||
| 8 | hO70ytxbBxukkFD60000 | None | Redun run report of 31972db9-19bb-4a1c-a7c5-41... | .txt | None | None | 6335 | ecV7smPo7CoZ1h59twt6Ow | None | None | md5 | True | False | 1 | 1 | None | None | True | NaN | 2025-06-10 20:15:08.580000+00:00 | 1 | None | 1 |
| 7 | FSUnzWN3cdRT1iXy0000 | None | log streams of run ZKnkCpA9XyR22zNM | .txt | __lamindb_run__ | None | 0 | 1B2M2Y8AsgTpgAmY7PhCfg | None | None | md5 | True | False | 1 | 1 | None | None | True | NaN | 2025-06-10 20:15:03.732000+00:00 | 1 | None | 1 |
| 6 | BbeoBehgpM00AlKF0000 | None | requirements.txt | .txt | __lamindb_run__ | None | 3202 | 7mCcF2ZlIp-m0c2vhuq4Mw | None | None | md5 | True | False | 1 | 1 | None | None | True | NaN | 2025-06-10 20:15:03.726000+00:00 | 1 | None | 1 |
| 5 | lQsh2ItUXoPEISoW0000 | data/results.tgz | None | .tgz | None | None | 83521 | DHxHjgueJGhZEU3DiM9WhA | None | None | md5 | False | False | 1 | 1 | None | None | True | 1.0 | 2025-06-10 20:15:03.671000+00:00 | 1 | None | 1 |
| 3 | DpeUKSeDgo5KxsYq0000 | fasta/KLF4.fasta | None | .fasta | None | None | 609 | LyuoYkWs4SgYcH7P7JLJtA | None | None | md5 | True | False | 1 | 1 | None | None | True | NaN | 2025-06-10 20:14:59.707000+00:00 | 1 | None | 1 |
| 4 | mdMdoSp7KpKk5EW60000 | fasta/SOX2.fasta | None | .fasta | None | None | 414 | C5q_yaFXGk4SAEpfdqBwnQ | None | None | md5 | True | False | 1 | 1 | None | None | True | NaN | 2025-06-10 20:14:59.707000+00:00 | 1 | None | 1 |
| 2 | UOFY7yU4ekvGM6i40000 | fasta/PO5F1.fasta | None | .fasta | None | None | 477 | -7iJgveFO9ia0wE1bqVu6g | None | None | md5 | True | False | 1 | 1 | None | None | True | NaN | 2025-06-10 20:14:59.706000+00:00 | 1 | None | 1 |
Feature
| uid | name | dtype | is_type | unit | description | array_rank | array_size | array_shape | proxy_dtype | synonyms | _expect_many | _curation | space_id | type_id | run_id | created_at | created_by_id | _aux | branch_id | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | ||||||||||||||||||||
| 1 | 3XaHn7J6Y5s4 | input_dir | str | None | None | None | 0 | 0 | None | None | None | None | None | 1 | None | None | 2025-06-10 20:14:56.864000+00:00 | 1 | {'af': {'0': None, '1': True, '2': False}} | 1 |
| 2 | otnFuI3fzi9E | amino_acid | str | None | None | None | 0 | 0 | None | None | None | None | None | 1 | None | None | 2025-06-10 20:14:56.864000+00:00 | 1 | {'af': {'0': None, '1': True, '2': False}} | 1 |
| 3 | v9M7GmBVwoWq | enzyme_regex | str | None | None | None | 0 | 0 | None | None | None | None | None | 1 | None | None | 2025-06-10 20:14:56.864000+00:00 | 1 | {'af': {'0': None, '1': True, '2': False}} | 1 |
| 4 | ZAaqJvrrhI1t | missed_cleavages | int | None | None | None | 0 | 0 | None | None | None | None | None | 1 | None | None | 2025-06-10 20:14:56.864000+00:00 | 1 | {'af': {'0': None, '1': True, '2': False}} | 1 |
| 5 | yLX4EIKQhVtu | min_length | int | None | None | None | 0 | 0 | None | None | None | None | None | 1 | None | None | 2025-06-10 20:14:56.864000+00:00 | 1 | {'af': {'0': None, '1': True, '2': False}} | 1 |
| 6 | 22SUV7J8xHhB | max_length | int | None | None | None | 0 | 0 | None | None | None | None | None | 1 | None | None | 2025-06-10 20:14:56.864000+00:00 | 1 | {'af': {'0': None, '1': True, '2': False}} | 1 |
| 7 | aK7kB6ch83SI | executor | str | None | None | None | 0 | 0 | None | None | None | None | None | 1 | None | None | 2025-06-10 20:14:56.864000+00:00 | 1 | {'af': {'0': None, '1': True, '2': False}} | 1 |
FeatureValue
| value | hash | space_id | feature_id | run_id | created_at | created_by_id | _aux | branch_id | |
|---|---|---|---|---|---|---|---|---|---|
| id | |||||||||
| 1 | ./fasta | G8diGhDfCu78R1WbXfsHOw | 1 | 1 | None | 2025-06-10 20:14:57.005000+00:00 | 1 | None | 1 |
| 2 | C | DWH4NwytHUEvgLhNFD4SVw | 1 | 2 | None | 2025-06-10 20:14:57.007000+00:00 | 1 | None | 1 |
| 3 | [KR] | zPuapwAPylpfWRQFOTw9Cg | 1 | 3 | None | 2025-06-10 20:14:57.009000+00:00 | 1 | None | 1 |
| 4 | 0 | z80ghJXVZe9m59_5-Ydk2g | 1 | 4 | None | 2025-06-10 20:14:57.011000+00:00 | 1 | None | 1 |
| 5 | 4 | qH_2eaLz5x2RgaZ7dUISLA | 1 | 5 | None | 2025-06-10 20:14:57.013000+00:00 | 1 | None | 1 |
| 6 | 75 | 0Jv0FUSjNlpGyQd-u141ww | 1 | 6 | None | 2025-06-10 20:14:57.015000+00:00 | 1 | None | 1 |
| 7 | Executor.default | BaNJiQQLvaFrvOtdglHU7w | 1 | 7 | None | 2025-06-10 20:14:57.017000+00:00 | 1 | None | 1 |
Run
| uid | name | started_at | finished_at | reference | reference_type | _is_consecutive | _status_code | space_id | transform_id | report_id | _logfile_id | environment_id | initiated_by_run_id | created_at | created_by_id | _aux | branch_id | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | ||||||||||||||||||
| 1 | ZKnkCpA9XyR22zNM | None | 2025-06-10 20:14:56.883967+00:00 | 2025-06-10 20:15:03.728786+00:00 | 31972db9-19bb-4a1c-a7c5-419180e73913 | redun_id | True | 0 | 1 | 1 | 8 | None | 6 | None | 2025-06-10 20:14:56.884000+00:00 | 1 | None | 1 |
Storage
| uid | root | description | type | region | instance_uid | space_id | run_id | created_at | created_by_id | _aux | branch_id | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | ||||||||||||
| 1 | QRS12R7Ta0Jk | /home/runner/work/redun-lamin/redun-lamin/docs... | None | local | None | iQlBPgD8uaqR | 1 | None | 2025-06-10 20:14:44.678000+00:00 | 1 | None | 1 |
Transform
| uid | key | description | type | source_code | hash | reference | reference_type | space_id | _template_id | version | is_latest | created_at | created_by_id | _aux | branch_id | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| id | ||||||||||||||||
| 1 | fncb7ZrLZP3V0000 | workflow.py | workflow.py | script | """workflow.py."""\n\n# This code is a copy fr... | mHyxrE622q2fluxICu4XDw | None | None | 1 | None | None | True | 2025-06-10 20:14:56.882000+00:00 | 1 | None | 1 |
Delete the test instance:
Show code cell content
!rm -rf test-redun-lamin
!lamin delete --force test-redun-lamin
• deleting instance testuser1/test-redun-lamin


